Uday Yallapragada and Iosif Vaisman
In this blog, I will summarize our results of network analysis of inter-protein correlated mutations in multiple Influenza A Virus (IAV) datasets. The methodology/pipeline that we conceived and realized are detailed here.
I will start with visualizations of these networks at 0.5 threshold. That is followed with plots of node counts and edge counts for different MIC threshold values, since this association can provide a general understanding of the degree of covariance. We also present MIC distribution plots since the network structure and dynamics is directly related to these distributions. Given our interest to understand the potential relationship between entropy and correlated mutation networks, I am also including details of average entropies for each of the proteins in all datasets. I have also created schematics of protein-level graphs for correlated mutation networks @ 0.5 MIC, to provide more obvious protein-level insights.
Network Visualizations
I created visualizations of correlated mutation networks at 0.5 threshold. I have included a static version of these network diagrams (for human H3N2, swine H3N2, human H1N1, swine H1N1 and avian H5 datasets) below.
These pictures clearly elucidate that there are significant differences in structural topologies of these networks. The density of nodes in human H1N1 network is significantly higher that swine H1N1 and the same comparison holds true for human H3N2 network over swine H3N2 network. The avian H5 network is dominated by residues from NA (purple) while the swine H1N1 network is characterized by four distinct clusters.
Node Counts
IAV strains that we downloaded from IRD consisted of a total of 4499 residues. We computed the total number of in-network residues for different MIC threshold values and noticed a wide variance based on the dataset. The number of nodes gradually decreases as MIC threshold increases. Several interesting observations can be made based on plots in figures shown below.
1. Human H1N1 vs Swine H1N1 (Figure 33) – The number of in-network residues for human H1N1 IAV correlated mutation network tends to be stable till a MIC threshold of ~0.6 after which it starts to decline while the number of in-network residues for swine H1N1 network gradually decreases for increasing values of MIC threshold. It should be noted here that the number of residues in human H1N1 network with MIC correlations in (0.1, 0.5) range is very low implying that we do not see new nodes joining the network for lower MIC values. While we see a slightly higher number of nodes in swine H1N1 at low threshold values in (0.1, 0.4) range, these numbers gradually decrease and the number of nodes with at least one significant mutation is lower compared to human H1N1 network. Both networks have similar node count for MIC values > 0.8.
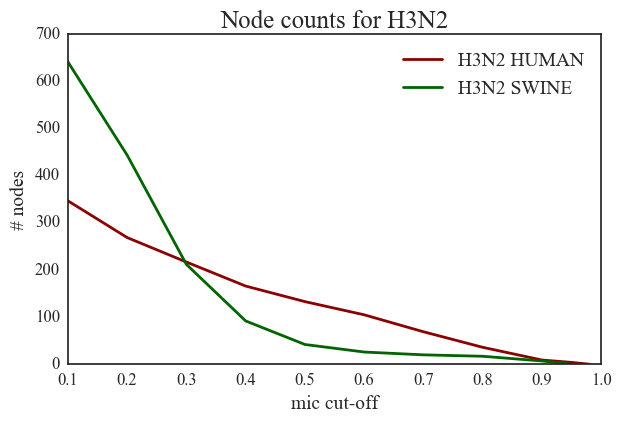
2. Human H3N2 vs Swine H3N2 (Figure 34) – We see a high number of in-network nodes (~650) for swine H3N2 strains at a low threshold value of 0.1 and we see a steeper decrease for increasing MIC threshold values and the number comes down to less than 50 for MIC threshold values in the significance region (>0.5). In other words, 98% of the residues in swine H3N2 strains do not have any significant correlated mutations. The human H3N2 network starts with a lower number of nodes (~350) at 0.1 MIC threshold and we see a much more gradual decrease in the number of nodes. We also observe a roughly overlapping tail for MIC > 0.88 in both these plots.
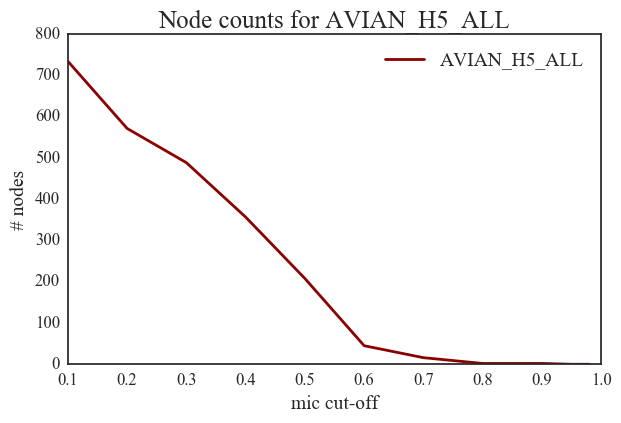
3. There are approximately 25 nodes in the H7N9 network in the significant zone (MIC > 0.5) while there are approximately 200 nodes in the significant zone for avian H5 network.


Edge Counts
The theoretical upper limit (max) for the total number of inter-protein edges in IAV strains approximates to 8.8 million. We computed the total number of edges in the network for different MIC threshold values. Plots for edge counts are depicted below.
1. We can make a general conclusion that less than 2% of edges in IAV have correlated mutations (MIC > 0.1) and a smaller fraction of edges have significant correlations.
2. There is a wide variance in edge counts based on the dataset.
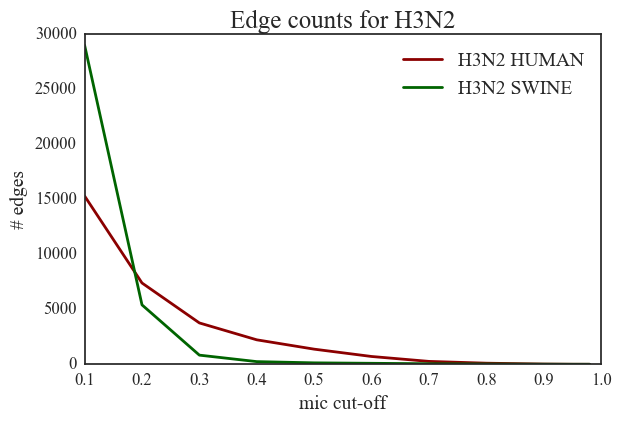
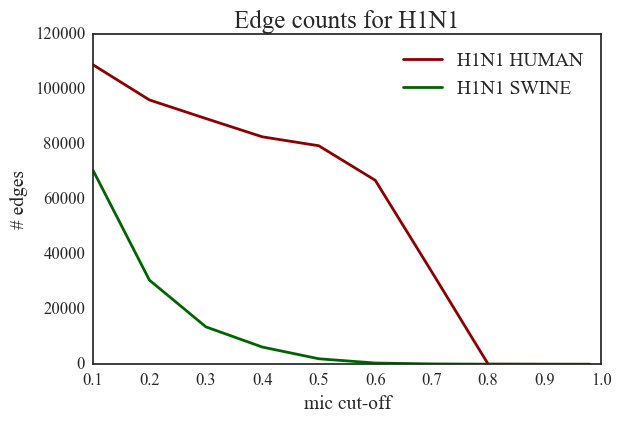
3. H1N1 vs. H3N2 – The number of edges in H3N2 networks is very small compared to edges in H1N1 networks.
4. Human H1N1 vs. Swine H1N1 – There is a significant difference between the number of edge counts for MIC > 0.5 in these two networks. There are 79524 significant edges in human H1N1 compared to a much smaller number (501) in swine H1N1.
5. Human H3N2 vs. Swine H3N2 – swine H3N2 network contains only 132 edges in the significant zone while the human H3N2 network contains 1378 significant edges. There is a steep decline in the number of edges (from 290000 to 5000) in swine H3N2 as the MIC threshold changes from 0.1 to 0.2.
6. We see more than 40000 edges in avian H5 for 0.1 MIC threshold but only 647 of these edges have MIC values greater than 0.5.
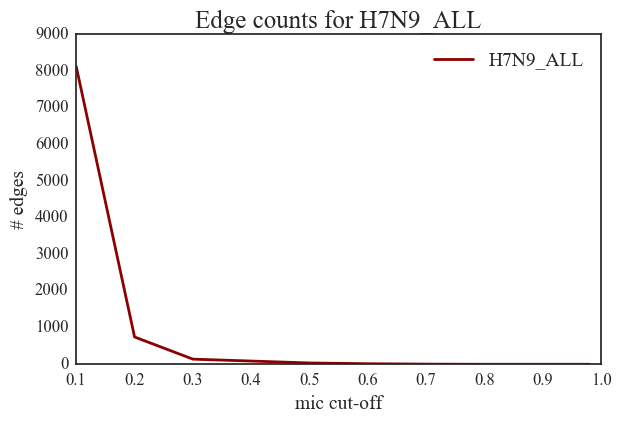
7. The H7N9 network is characterized by a very small number of edges. This network contains only 37 edges with MIC values greater than 0.5.
8. Like our observation with ‘node counts’, the number of edges in ‘Human All’ network is significantly higher compared to all other datasets.

Degree Distribution
The degree of a vertex is the number of edges emanating from it. Degree distribution is an important characteristic of a graph and provides a distribution of the degree of nodes over the entire network. Degree distribution reflects the overall pattern of connections in a dataset. A node with high degree in correlated mutation network implies that the residue has correlated mutations with many residues in the network.
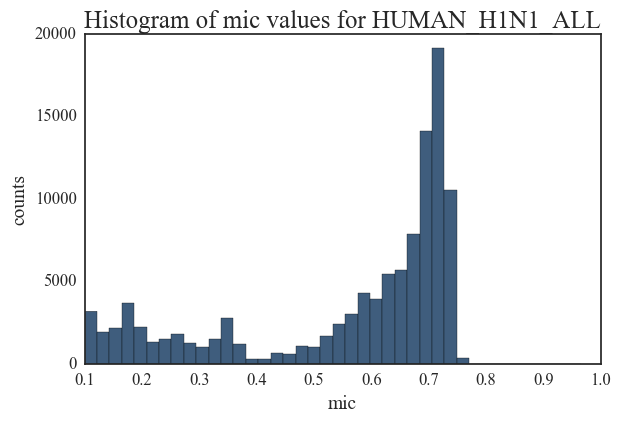
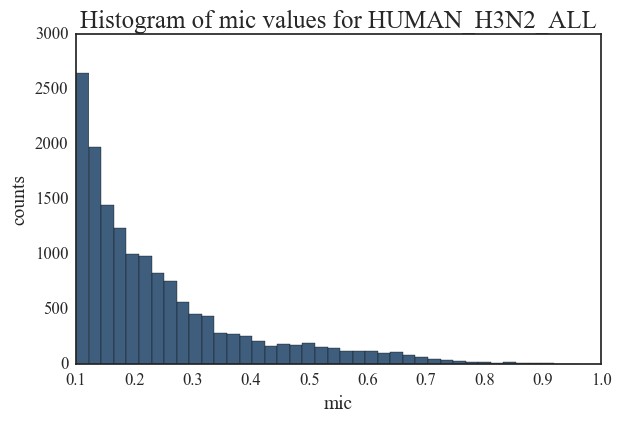
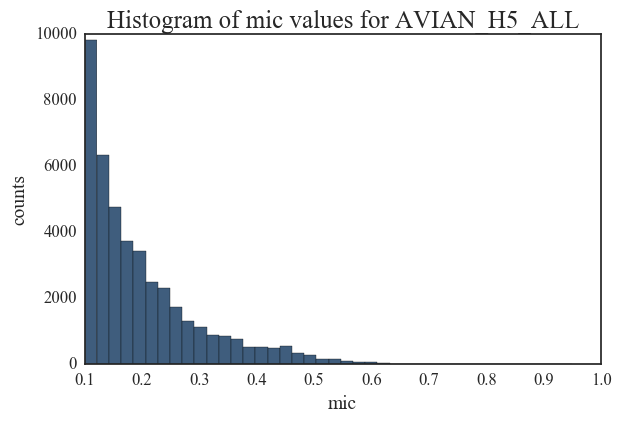
I am including histograms of degree distributions here. There is substantial difference between each of these degree distributions indicating that the overall structure of a correlated mutation network in IAV does not adhere to a single topology. While the distributions for Swine H1N1 and Avian H5 are close to a power-law model, other network distributions are more indicative of random networks. These plots illustrate the complex evolutionary patterns in Influenza and highlight the fact that the overall mutation profile and evolution in IAV strains are host and sub-type specific.

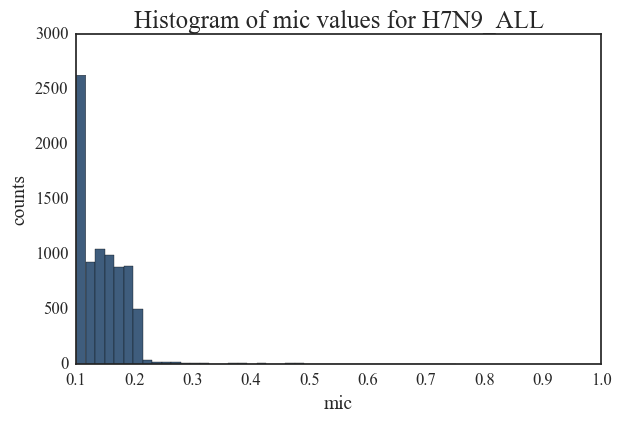
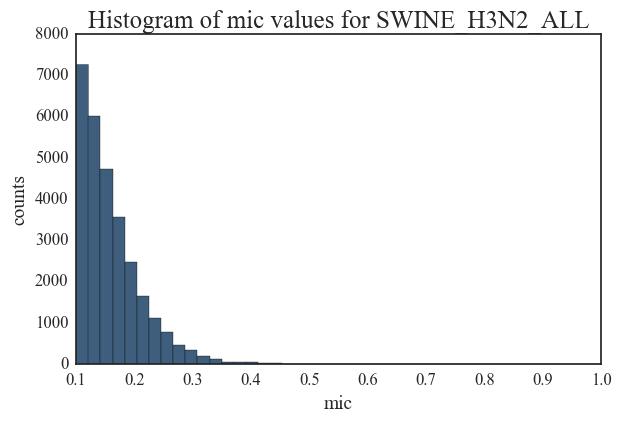
Entropy
To gain a global view of sequence variation, we calculated average entropy values for sequences of all the 10 proteins in different datasets. This average entropy plot (see below) revealed that NA protein in Avian H5 has the highest overall sequence variation. Proteins in Human-All, Avian H5, Swine H1N1 and Swine H3N2 datasets had higher entropies compared to proteins in H7N9, Human H1N1 and Human H3N2 data sets. Within each dataset, HA, NA and NS1 had the highest average entropy among all the proteins.
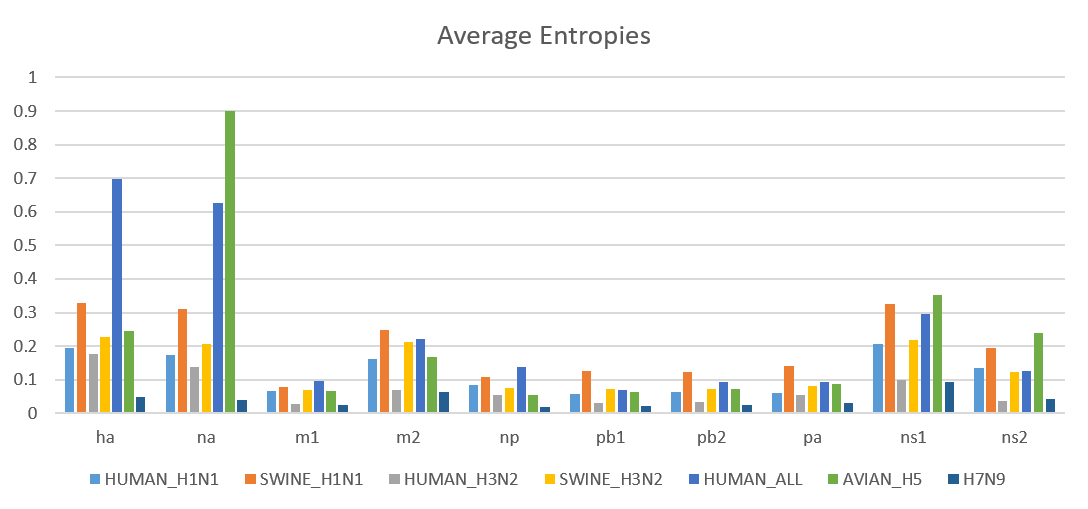
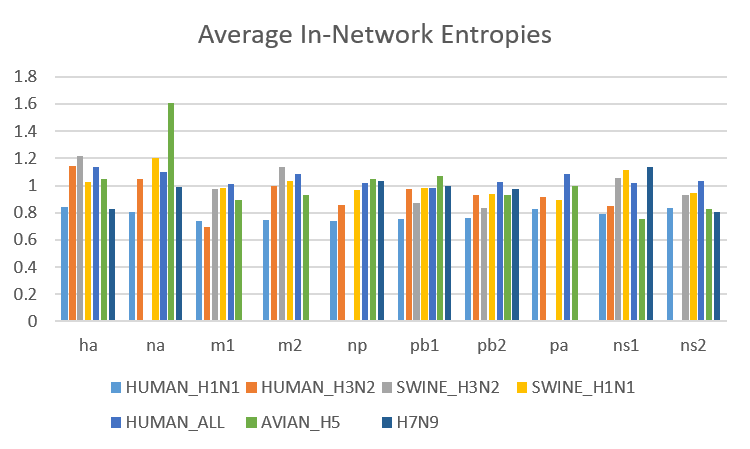
We have also created separate plots of average entropies for in-network and out-of-network residues. These plots revealed that the entropy values of in-network residues are generally higher than the entropy values of out-of-network residues.

Protein Correlation Graphs
To understand the extent of co-variation between the 10 proteins in IAV sequences, we created visualizations of protein correlation graphs where the proteins acts as nodes and connections between these nodes are derived based on correlated mutations between residues. Strength of a node (enumerated in parenthesis as part of the name of the node) is the total number of residues in that protein with at least one significant correlation with a residue in another protein, while the strength of a connection (depicted by the thickness of a connection) is the total number of edges between residues in two proteins. Several interesting observations can be made from these visualizations.
1. HA and NA proteins play the most prominent role in protein correlation networks. They tend to have the maximum number of residues with significant correlations.
2. NA protein dominates the Avian H5 network (Figure 73).
3. Figure 71 and Figure 72 elucidate the differences between Human H3N2 and Swine H3N2 networks. The Swine H3N2 network is sparse and contains very few connections between proteins compared to the Human H3N2 network.
4. Protein interaction networks of Human H1N1 and Human H3N2 (Figure 70, Figure 73) suggest that residues in NP have the third highest number of residues with correlated mutations (after HA and NA).
5. These networks (except for H7N9 network) contain residues from all 10 proteins.







