In this blog, I provide a quick overview of ‘correlated mutations’ in proteins.
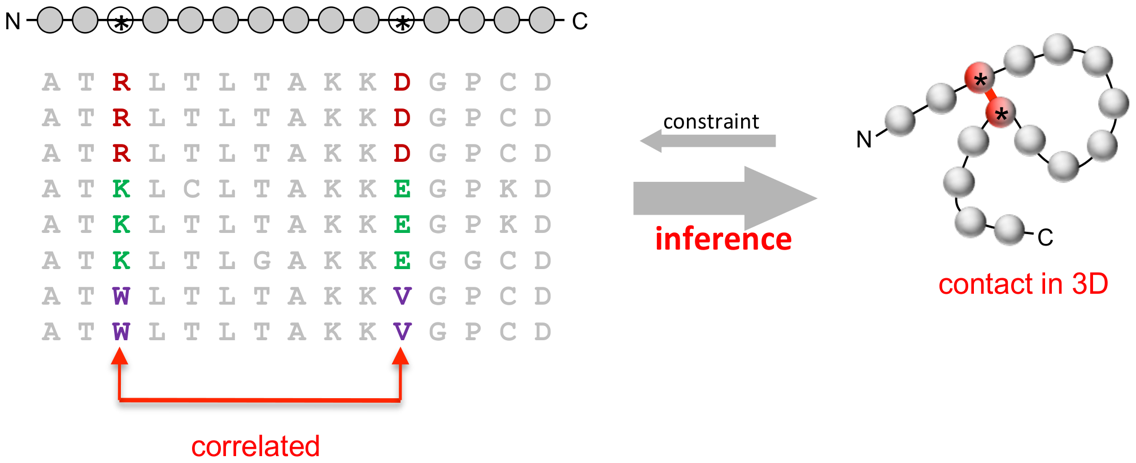
What exactly is a correlated mutation? Let’s take a look at the following picture to understand correlated mutations among positions within a single protein (or intra-protein correlated mutations).
 [1]
[1]
This picture depicts a multiple sequence alignment of residues in a segment of a hypothetical protein. Each of these sequences is from from a separate sample/strain. The changes in position #3 and position #11 in the above picture are called correlated mutations since amino acids at these positions are covarying . More specifically, when R (Arginine) changes to K (Lysine), D (Aspartic acid) changes to E (Glutamic acid) and when R changes to W (Tryptophan), D changes to V (Valine).
These ‘correlated mutations’ can also occur between residues in two different proteins (inter-protein correlated mutations). The following picture depicts ‘correlated mutations’ at two sites in 2 different proteins.

The term “correlated mutation” with a protein context was formally introduced in [2] as ‘tendency of positions in proteins to mutate coordinately’. Sequence correlation/covariance analysis is an area that has gained significant traction over the last decade and is widely used for identifying correlated sites in proteins. In this context, it is important to note that correlated mutations between residues in proteins have initially been linked primarily to probable physical contact in three-dimensional space but more recent studies have demonstrated that co-mutation of amino acids may originate not only from structural contacts but also from a much broader range of biological reasons. More specifically, studies in this field have suggested that
(a) correlated mutations may also occur due to reasons related to protein function [3]
(b) coevolution between amino acid residues is necessary and sufficient to specify sequences that fold into native structures [4]
(c) and residues highly correlated with others are indeed more likely to be associated with disease [5].
References
- PLOS One, Protein 3D Structure Computed from Evolutionary Sequence Variation, Dec 2011
- D. Altschuh, A. M. Lesk, A. C. Bloomer, and A. Klug, “Correlation of co-ordinated amino acid substitutions with function in viruses related to tobacco mosaic virus,” J. Mol. Biol., vol. 193, no. 4, pp. 693–707, Feb. 1987.
- I. N. Shindyalov, N. A. Kolchanov, and C. Sander, “Can three-dimensional contacts in protein structures be predicted by analysis of correlated mutations?,” Protein Eng. Des. Sel., vol. 7, no. 3, pp. 349–358, Mar. 1994.
- U. Göbel, C. Sander, R. Schneider, and A. Valencia, “Correlated mutations and residue contacts in proteins,” Proteins Struct. Funct. Bioinforma., vol. 18, no. 4, pp. 309–317, Apr. 1994.
- G. B. Gloor, L. C. Martin, L. M. Wahl, and S. D. Dunn, “Mutual Information in Protein Multiple Sequence Alignments Reveals Two Classes of Coevolving Positions,” Biochemistry (Mosc.), vol. 44, no. 19, pp. 7156–7165, May 2005.
